|
Description of biowep, a Web Enactment Portal for Bioinformatics.
biowep is a web-based client application that allows for the selection and execution of a set of predefined, annotated workflows involving
access to and retrieval from bioinformatics tools, either databases or software tools.
Although biowep is intended to be a general tool, presently it can only include workflows developed by using either the
Taverna Workbench or BioWMS (see
Bartocci E et al. BioWMS: a web-based Workflow Management System for bioinformatics. BMC Bioinformatics 2007 8(Suppl 1):S2.
DOI: 10.1186/1471-2105-8-S1-S2).
biowep is based on:
-
a Workflow Manager (WM), for the creation and annotation of workflows. Input and output data, elaboration type and
application domain of main steps of each workflow are annotated by using a classification of bioinformatics data and tasks.
-
a User Interface (UI), for authentication and profiling of end users, selection of workflows from lists or by searching their
annotation, management of results.
-
a Workflow Executor (WE), for controlling the enactment of workflows that are carried out either by
FreeFluo
or by Hermes, a mobile agent-based middleware.
The overall architecture scheme is shown in the following figure (click on it to see a larger version).
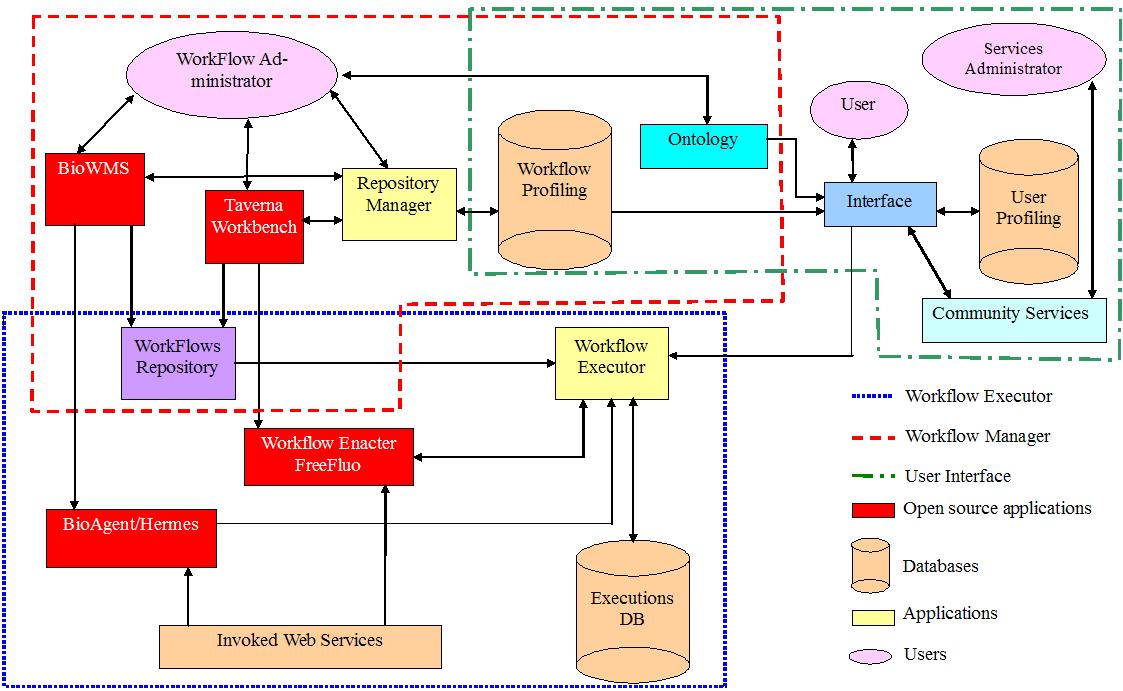 .
biowep is partially based on open source and it is itself available under the GNU Lesser General Public Licence (LGPL).
biowep is derived from the prototype developed by the Oncology over Internet
project and it was further developed in the sphere of the Laboratory of Interdisciplinary Technologies in Bioinformatics project.
These pages are available for researchers interested in the development of biowep.
The biowep portal is not presently maintained.
|