Combining Searches
Introduction
General Syntax
(See section 8.3.3 "Operands" and section 8.3.4 "Operators" for more information on operands and operators, respectively.)
For example:
where enzyme and pdb are operands specifying the databanks, Enzyme and PDB respectively, and > is an operator telling SRS to search for links between the databanks, and keep only those entries which belong to the databank on the right. The result of this query is a list of entries in the PDB databank that have links to the Enzyme databank.
Combinations may also include index searches. For instance:
will create a list of all the entries in the PDB databank that have links to the results of the index search:
See section 8.2 "Searching in Indices" , for an explanation of this sub-search.
The above examples are fairly trivial, but the SRS query language allows you to build up more complex queries using the various operators. In addition expressions can be grouped using parentheses so that they are treated as a single entity. You will see examples of searches and ways of combining them, throughout this chapter.
Operands
| Databank name | EMBL | Each databank has a unique name. |
| Set name | Q1 | SRS gives each query a name which can be used when you want to perform an operation on the set of results from that query. |
| Index search | [embl-des: kinase] | This command initiates a search in one or more indices, of one or more databanks. |
| Expression | (Q1&Q2) | If an expression is enclosed in parentheses, it is treated as a single operand. Parentheses can be nested to any degree. |
| Parent | parent | This is a special operand that allows the conversion of a set of subentries into a set of entries (see section 8.4 "Entries and Subentries"). |
Operators
Logical Operators
The logical operators, OR (|), AND (&) and BUTNOT (!), can be used to combine search words in an index search, or to combine sets in a query.
The following figure illustrates the effects of the three operators in an expression of the form, A operator B.
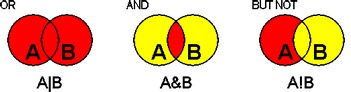
Figure 8.2 Logical operators in SRS.Logical operations can only be performed between sets of the same type. It is not possible, for instance, to combine a set of entries and a set of subentries (see section 8.4 "Entries and Subentries") using logical operators. In such cases, an additional link operation must be specified (see section 8.4.1 "Links with Sets Containing Subentries").
Link Operators
Link operators are unique to the SRS query language. The two link operators, < and >, allow sets of data from different databanks to be combined.
Figure 8.3 shows two databanks, A and B, in which some entries in A have links to entries in B. These links are processed to build link indices that provide the basis for the link operation. The figure shows the results of two searches for links between sets A and B, using the operators, < and >.
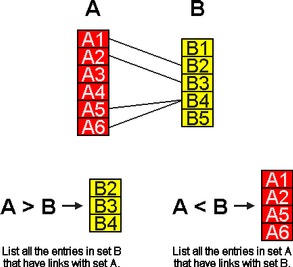
Figure 8.3 Linked Data.Links are not usually bidirectional; however, the link indices in SRS are used bidirectionally. For instance:
- A > B
- This retrieves those entries in B that are linked to entries in A.
- A < B
- This retrieves those entries in A that are linked to entries in B.
Use of Operators to Combine Search Items
This example searches for links to a specified databank in the results of an index search:
The result will be a list of all the entries in the PDB databank that have links to the results of the index search:
See section 8.2 "Searching in Indices" , for an explanation of index searches.
Example 8.3 Multiple linking 1
It is possible to combine several linking queries. For example:
The search first retrieves the SWISS-PROT entry "acha_human". This entry will have links to PROSITE entries that document the protein family of which "acha_human" is a member. (In this case it is the family of neuronal acetylcholine receptors.) These items in the PROSITE databank are retrieved. The next link retrieves SWISS-PROT entries that are linked to the PROSITE entries, i.e., belong to the neuronal acetylcholine receptor family. In effect, the entry "acha_human" is being amplified to retrieve all the entries in SWISS-PROT which document members of the protein family or families to which it belongs.
Example 8.4 Multiple linking 2
A similar technique to that used in example Multiple linking 1 , can be used to find related information in another databank to which the initial entry is not linked.
The query retrieves a probable glutathione reductase (whose ID is "gshr_caeel") from SWISS-PROT, searches for entries in ProDom which document related proteins, and then looks for links to PDB in these entries. The result is a set of PDB protein structures that are homologous to the SWISS-PROT entry "gshr_caeel".
This example can be thought of as being composed of two parts:
The first part of the query searches the description fields of the SWISS-PROT and SWISSNEW databanks, looking for "kinase". The second part of the search excludes any entries that have links to the SWISSNEW databank. Using this technique, it is possible to retrieve the entries for "kinase", but exclude any that are replaced by (more up to date) entries in SWISSNEW.
(q = [{swissprot swissnew}-des:kinase])!(q<swissnew)
The distinction being made here is that entries in SWISSNEW will not be linked to themselves (or other entries in SWISSNEW), so all the SWISSNEW entries will be kept. However, any entry in SWISS-PROT that has been replaced by an entry (to which that entry is linked) in SWISSNEW will be picked up and rejected. In this way, out-of-date entries are excluded.
Note:The first part of the query also defines a group, q, which contains the SWISS-PROT and SWISSNEW databanks. This is used in the second part of the query rather than listing the databanks explicitly (see section 8.2.6 "Searching Multiple Databanks").
Example 8.6 Searching multiple databanks and screening for overlaps
Many protein or DNA databanks overlap to a great extent, which creates a lot of redundancy; however, the annotation of equivalent entries in different databanks can be quite varied. This can be useful for string searching because the probability of finding a certain enzyme name is greater if you can search all sequence databanks. After the search, links can be used to remove any overlaps.
See Example Complex linking , for how this might be done.
