|
Welcome to help page on CLIMA 2, the Cell Line Integrated Molecular Authentication database, version 2.
This system is under active development, and help pages are also developing accordingly.
Please forgive us for possible errors and send us your comments, criticisms and congratulations, if any.
Citing CLIMA 2
If you use CLIMA 2, please cite the following reference:
P. Romano, A. Manniello, O. Aresu, M. Armento, M. Cesaro, B. Parodi.
Cell Line Data Base: structure and recent improvements towards molecular authentication of human cell lines.
Nucleic Acids Research 2009 37(Database issue):D925-D932.
DOI: 10.1093/nar/gkn730;
PMID: 18927105
Overview
CLIMA 2 is the result of an integration effort aimed at including all certified STR profiles of human cell lines in a unique database.
The contents of the database are dinamically summarized (i.e., they are always up-to-date) in a table in the home page.
Using the search engine should be straightforward and results are displayed in a simple format.
Nevertheless, what follows is a quick guide on how-to search CLIMA 2 and understand results.
You can use CLIMA 2 to identify your cell lines.
Moreover, You can search CLIMA 2 database either by name or by locus.
Results are displayed by using similar formats:
see identification result page, results page of the search by name
and results page of the search by locus.
Query forms also include some fields for statistical aims, i.e. email address and country of the user.
You are welcome to fill them or not.
If you fill in the email address field with a valid email address, you will receive a summary of your query by email.
Identification of cell lines
The identification of a cell line is performed according to the standard
ANSI/ATCC ASN-0002-2011 on "Authentication of Human Cell Lines: Standardization of STR Profiling".
See ANSI eStandards Store.
This standard makes reference to the XXXXXX STR kit from YYYYY.
This kit includes the following STR loci: D5S818, D13S317, D7S820, D16S539, VWA, TH01, Amelogenin, TPOX, CSF1PO.
In this context, the standard states that:
"A cell line is considered misidentified when ≥ 80% of the alleles in its STR profile match a cell line in the database from a different donor.
Similarly, a cell line is considered misidentifed if it fails to match tissue or otehr cell line samples known to be come from the same donor."
The identification tool in CLIMA 2 allows users to submit the STR profile of a cell line they want to identify.
For each locus, up to four distinct values can be entered, but for the Amelogenin, for which only up to two values (X and Y) can be entered.
These values can be entered in any order, but must be all different. In other words, they can't be repeated.
The desired lower percent match to show must also be entered.
The STR profile is checked against all profiles in CLIMA 2 determined by using a kit having the same loci.
The number of matches, i.e. of identical values for the same locus, is computed for the entered profile and each profile in the database.
For each pair, the Standard Percent Match (StPM), that is the ratio between the number of matches and the number of distinct values in the profile in the database, is then computed.
|
|
|
| Figure 1: cell line identification input form |
|
Identification result page
The result page of the identification tool reports the list of all STR profiles in the database that have a StPM with the profile entered by the user
that is greater then or equal to the lower percent match value entered by the user in the query form.
The list is ordered by decreasing percent match value. Profiles having the same percent match, are ordered by decreasing number of loci values in their profiles
and, in case of further equality, by name in alphabetical order.
NB! Only those profiles that have the standard set of loci are queried.
However, all loci included in the profile are shown, even when they were not accounted for the query.
The results page includes:
- A short summary of the search (list of all loci involved in the search with submitted values and the lower percent match).
- A table including STR profiles of cell lines with a percent match greater then or equal to to given threshold.
The first row of the table reports the queried profile.
Each following row in the table includes:
- the name of the cell line, the dataset it belongs to, and the catalogue code of the line, when it exists
- the percent match with the queried profile, computed according to the standard as explained above
- the STR profile values (first the loci of the standard kit, then all remaining loci)
NB! Values of the database profiles are shown in red when they match the query, in black otherwise.
|
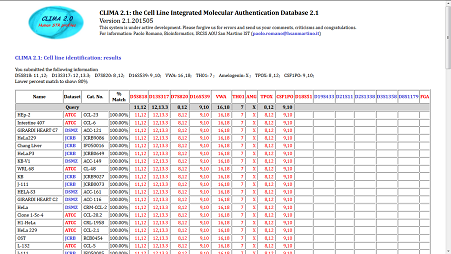
|
Figure 2: example of the identification result page
(click to open in a new tab and enlarge) |
|
Search by name
Search by name looks in the database for all STR data that are assigned to cell lines whose name matches the submitted term.
Some simple rules apply to the submitted string:
- letter cases do not care: "HeLa", "hela" and "HELA" are the same string for the system.
- spaces are included in the submitted string: "HeLa 229" is a valid name, the blank space is included in the search.
"HeLa " is also a valid search value, including the final " " (this can be used, e.g. as a prefix in searches (see below).
- any character included in the string is used in the search, but in the specific case: "MRC-5" is different from "MRC5" and
if either single or double quotes are inserted into the search field, they are indeed included in the searched name.
While searching by name, you can choose to supply an exact name, a prefix or a string that will be evaluated "by sound".
- Exact name: the name is searched against the database for cell lines whose name is identical to the submitted string.
Examples:
- Search: "HeLa"; Look for: "HeLa"; Cannot find: "HeLa 229"
- Search: "MRC-5"; Look for: "MRC-5"; Cannot find: "MRC5"
- Truncated name: the name is searched against the database for cell lines whose name begins by the submitted string.
Examples:
- Search: "HeLa"; Look for: "HeLa%"; Find everything that begins by "HeLa", like "HeLa 229", "HeLa229", "HeLa.P3"
- Search: "HeLa "; Look for: "HeLa %";
Find everything that begins by "HeLa " (including the blank space), like "HeLa 229" and "HeLa S3";
Cannot find: "HeLa" alone;
Cannot find names including "HeLa ", but not beginning by "HeLa ".
- By sound name: the name is searched against the database for cell lines whose name has the same (or a very similar)
sound of the submitted string. Here, the special function soundex() is used. This function is fequently used in database
systems. It computes a key (the soundex key) that is not unique for the generating word. Instead, all words pronounced
similarly produce the same key, and can thus be used to simplify searches in databases where you know the pronunciation,
but not the spelling. The soundex function returns a string 4 characters long, starting with a letter and it
therefore normally only reflect the initial part of the word.
This particular soundex function is one described by Donald Knuth in "The Art Of Computer Programming, vol. 3:
Sorting And Searching", Addison-Wesley (1973), pp. 391-392.
Examples:
- Search: "HiLa"; Look for: soundex("HiLa");
Find everything that has a similar sound like "HeLa", "HeLa 229", "HEL-11", "HLE", "HLE"
- Search: "Molt", Melt or "Malt"; Look for: soundex("Molt") or sounex("Melt") or soundex("Malt");
Find everything that has a similar sound, including "MOLT-4".
|
Search by locus
Search by locus looks in the database for all STR data corresponding to the searched loci values.
There is one row of four values for each locus that is included in at least one of the datasets that
are available in the CLIMA 2 database.
NB! As a consequence, a search can be created for a set of loci that are not all present in the same dataset.
The following rules apply:
- At least one value must be inserted for at least one locus.
The search is performed by using the leftmost value of each locus to determine if the locus is
involved in the search. All loci for which none value is specified are not used in the search.
- Logical operations are first executed for each locus and are then combined together by a logical AND.
This means that cell lines must comply with conditions applied to all loci.
There is an important exception to this rule: since the user can insert conditions on loci
that are not available for all datasets, searches on single datasets are performed only by using
those loci that are included in the dataset.
Examples:
- Search: 17 in locus D18S51 and Y in locus Amelogenin. Amelogenin is included in all datasets,
while D18S51 is only included in some.
Search: values for D18S51 and Amelogenin are used for
searching those datasets that have both loci, Amelogenin alone is used for searching those
datasets not including D18S51.
- Only one value can be included in each field. If it is a decimal number, the dot (".") must be used as
a separator. Single and double quotes must not be inserted.
Examples:
- Search: "10,12"; Find: nothing.
- Search: "10.1"; Find: all cell lines that have the value 10.1 in the specified locus,
either alone or with any other value.
- If only one value is included for one locus,
it must be inserted in the leftmost field.
- If only one value is included for one locus,
the system looks for all cell lines that has the specified value,
either alone or with any other value.
- If more than one value is included for one locus,
the leftmost field must be used anyway and the other values must be inserted into adjacent fields, left to right.
- If more than one value is included for one locus, the order is not influent on the result.
- If more than one value is included for one locus,
they are combined according to the logical operator (AND or OR) specified between them.
The logical AND has precedence over the logical OR.
This implies that all AND operations are perfomred before any OR operation can accour.
Examples:
- Search: values 17 and 18 for locus D18S51, operator AND.
Search expression: (D18S51=17 AND D18S51=18) NB! This is not strange, D18S51 may have up to four different values...
Find: all cell lines having both 17 and 18 for D18S51, including those that have further values,
like e.g. 13,17,18 and 16,17,18.
- Search: values 13, 17 and 18 for locus D18S51, operators OR and AND.
Search expression: (D18S51=13 OR (D18S51=17 AND D18S51=18))
Find: all cell lines having both 17 and 18 for D18S51, including those that have further values,
and all cell lines having 13 for D18S51, including those that have further values.
E.g., the following sets are all found: (13), (17,18), (16,17,18), (13,17), (13,16).
- There is an exception to the previous rule for Amelogenin. Since this locus can only be "X" or "X,Y"
(never empty or "Y" alone), the logical operator is obligatory an AND.
|
Results page for the search by name
The search by name provides data for those cell lines whose name match the query and
all STR data available for them in the datasets included in the CLIMA 2 system.
The results page includes:
- A short summary of the search that is based on the searched name and the searching criteria.
- The list of all cell line names and, for each of them, the indication of datasets where they are available.
- A table for each dataset including loci values and all information known about the selected cell lines.
The list of cell line names with indication of data by available datasets is displayed by using a table.
The first column includes cell line names, the following columns (one for each dataset) specify whether
the cell line name is included in the dataset (in which case, a 'Yes' is displayed) or not ('No).
The headings of columns that relate to a dataset consist in the dataset name.
A link to the original dataset from where the information was taken is connected to the name.
The following tables, one for each dataset, have similar formats, although information can differ.
The table is preceded by a short title where a link to the on-line original information is connected to the
name of the dataset.
In general, the first column refers to the cell line name and the table includes at least one further column
for each locus in the dataset.
The headings of columns referring to loci values consist in the name of the locus.
A link to the description of the locus provided by the "Short Tandem Repeat DNA Internet DataBase (STRBase)"
is connected to the name.
NB! A paper on STRBase was published in
Nucleic Acids Research Molecular Biology Databases Supplement in 2001.
See the paper and the
Supplement on-line.
Additional columns can be provided, depending on the dataset.
If the dataset includes the cell line code in the collection catalogue, this is listed and, if applicable, a link
to the cell line's description is connected to the name.
|
Results page for the search by locus
The search by locus provides data for those cell lines that match submitted loci values for all
datasets for which this information is applicable.
NB! Datasets are searched only on the basis of the loci that they include.
If the search included a set of loci that are partially available in a dataset, it will
be searched by using only that subset. This can happen, e.g., when Amelogenin is involved
in the search, since this locus is present in all datasets.
The results page includes:
- A short summary of the search that is based on the list of all loci involved in the search with submitted values.
- A list of hits number for each dataset
- A table for each dataset including loci values and all information known about the selected cell lines.
Datasets' tables have the same structure shown in the results page for the search by name.
|
For information, get in touch with:
Paolo Romano, Bioinformatics, IRCSS Ospedale Policlinico San Martino
Email: gabriela.coronel@hsanmartino.it
|
 |
If you use CLIMA 2, please cite the following reference:
Romano P, Manniello MA, Aresu O, Armento M, Cesaro M, Parodi B.
Cell Line Data Base: structure and recent improvements towards molecular authentication of human cell lines.
Nucl Acids Res 2009 37(Database issue):D925-D932.
DOI: doi:10.1093/nar/gkn730
|
|


